Source at GitHub: https://github.com/iprokin/Py_XPPCALL
Introduction
XPPAUT is a great software for the analysis of dynamical systems. Many mathematical models implemented with XPPAUT (*.ODE file format) [search for examples at http://senselab.med.yale.edu/modeldb/]. These implementations could be used to reproduce computational experiments and quickly test ideas. For instance, one could perform sensitivity analysis of model parameters or search parameter space. However, due to the graphical nature of XPPAUT, a batch modification of model parameters is problematic.
Here I describe the solution to this problem using a simple python module Py_XPPCALL that I have written to interface with XPPAUT. It allows you to batch-modify model parameters and returns corresponding solutions. A similar software exists https://github.com/jsnowacki/xppy, however, little documentation is available.
Py_XPPCALL is easy to use and it does not require installation. Simply put xppcall.py to your python path or current folder.
Numerical optimization example with Hodgkin-Huxley neuron model
I extracted the ODE source code of Hodgkin-Huxley model from the example at the website of XPPAUT http://www.math.pitt.edu/~bard/bardware/tut/newstyle.html#hh. I saved it as hh.ode and I put it to the same folder where I have xppcall.py and ran python from there.
## Py_XPPCALL Example
# import some modules including Py_XPPCALL
import matplotlib.pylab as plt
import numpy as np
from xppcall import xpprun, read_pars_values_from_file
# Let's check what are the parameters of the model
pars = read_pars_values_from_file('hh.ode')
print pars
Out[]:
{'c': '1',
'gk': '36',
'gl': '.3',
'gna': '120',
'i': '0',
'vk': '-77',
'vl': '-54.4',
'vna': '50'}
# Note: XPPAUT is not case sensitive. In Py_XPPCALL, the names of parameters and variables were chosen to be in lower case.
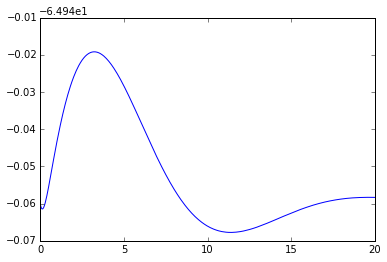
# Let's plot solution for membrane potential with parameters specified in .ODE file
npa, vn = xpprun('hh.ode', clean_after=True)
plt.figure()
plt.plot(npa[:,0], npa[:, 1+vn.index('v')])
Out[]:
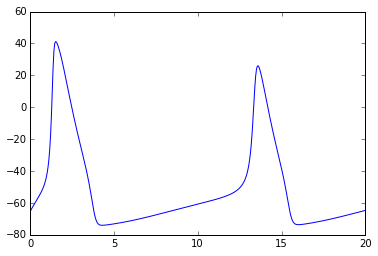
# Let's modify constant input current
npa, vn = xpprun('hh.ode', parameters={'i':20.0}, clean_after=True)
plt.figure()
plt.plot(npa[:,0], npa[:, 1+vn.index('v')])
Out[]:
# Example of an optimization using fmin from SciPy
from scipy.optimize import fmin
# define desired V graph
target_v = -80.0+20*npa[:,0]*(np.sign(-npa[:,0]+4)+1)
plt.figure()
plt.plot(npa[:,0], target_v)
Out[]:
# define objective_func we want to minimize
def objective_func(x, target=target_v):
try:
npa, vn = xpprun('hh.ode', parameters={'i':x[0]}, clean_after=True)
computed = npa[:, 1+vn.index('v')]
return np.mean( (computed-target)**2 )
except:
return 1e10 # return huge error if something went wrong
xopt = fmin(objective_func, [20.0]) # find new parameters
npa, vn = xpprun('hh.ode', parameters={'i':xopt[0]}, clean_after=True)
plt.figure()
plt.plot(npa[:,0], npa[:, 1+vn.index('v')], label='fit')
plt.plot(npa[:,0], target_v, label='target')
plt.legend()
Out[]: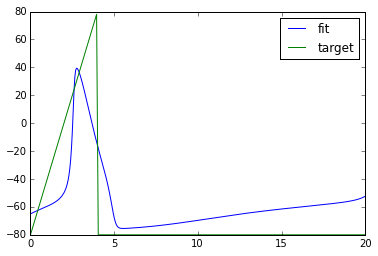
Congrats! We got a pretty good fit.
How to Cite Py_XPPCALL
If you want to refer to Py_XPPCALL in a publication, you can use
“Prokin, I. and Park, Y. Py_XPPCALL. 2017. Available at: https://github.com/iprokin/Py_XPPCALL.”
or BibTeX:
@misc{xppy, author = {Prokin, Ilya and Park, Youngmin}, title = {{Py_XPPCALL}}, url = {https://github.com/iprokin/Py_XPPCALL}, year = {2017} }